With the emergence of the Omicron variant of concern (VOC) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent that causes the coronavirus disease 2019 (COVID-19), new and accurate diagnostic tests are needed to distinguish infection with this strain from that caused by other variants. A new study published on the preprint server medRxiv* reports a method to do this using four polymerase chain reaction (PCR) tests.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
The SARS-CoV-2 Omicron variant was first described in South Africa and Botswana in November 2021. Soon after, the Omicron variant was detected in many other countries including Hong Kong and Israel.
This variant has 30 mutations in its spike protein, making it the most highly mutated variant of SARS-CoV-2 to date. Of these mutations, 15 affect the receptor-binding domain (RBD), which is a component of the spike protein that makes contact with the angiotensin-converting enzyme 2 (ACE2) receptor on host cells.
Most diagnostic tests today focus on the use of reverse transcription-polymerase chain reaction (RT-PCR) to amplify minute amounts of viral nucleic acid in the sample and thus make it possible to identify its presence and type. Using four different assays of this type the researchers in this study were able to carry out a quick identification of the new variant B.1.1.529.
The assays do this by amplifying and then binding to characteristic mutations in this variant, namely, in the non-structural protein nsp6, the viral spike, and the nucleocapsid genes. Not only are these assays simple to perform, but they are also suitable for multiplexing with a little adjustment and can be used to detect the presence of the Omicron VOC in routine test samples. This preliminary report will need to be followed up and the findings validated.
The need to find a way to quickly identify the Omicron variant spurred the development of the four assays described here by the Israel Ministry of Health Central Virology Laboratory (CVL) and the Israel Institute for Biological Research (IIBR). Based partly on existing protocols, this allows mutations already firmly linked to this variant to be detected using sequences uploaded to the Global Initiative for Sharing Avian Influenza Data (GISAID) database.
The assays were tested against the earliest Omicron samples in Israel and compared against the Delta variant which was, at that time, the only strain of SARS-CoV-2 in circulation in Israel. A few Alpha- and wild-type strains were also used to test for specificity of assay performance.
The test was modified to allow multiplexed testing based on established reactions already in use by the United States Centers for Disease Control and Prevention (CDC).
“The assays can be implemented in any molecular diagnostic laboratory and do not require the use of specific brand instruments, or unique reagents.”
The mutations which these assays are designed to detect include deletion of spike gene positions 22194-22196 and nucleocapsid gene deletion 28,362-28,371 since these are found only in the Omicron variant. These were found in all Omicron sequences uploaded so far.
Negative Ndel reaction
The assay picks up N deletions 28362 using the N1 R1 reverse primer to ensure selectivity. When this deletion is present, the primer annealing is strongly inhibited 104-fold, thereby leading to the absence of any signal or a 12-15 difference in Cq values for the E-sarbeco reaction and the Ndel reaction. The former includes the deleted version and the latter does not.
By making use of the readily available primer and probe developed by the CDC, the researchers ensured the test can be easily performed in all laboratories. The assay targets the deletion at the position of reference sequence NC_045512.
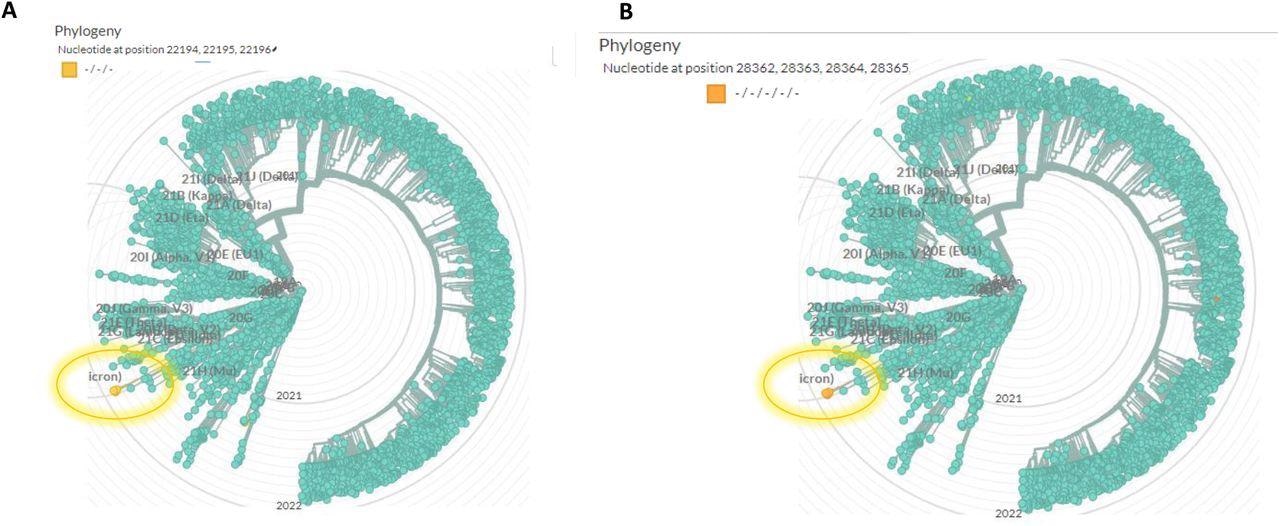 Global phylogenetic analysis shows the uniqueness of the spike 22194-22196 deletion (A) and the nucleocapsid (N) 28362-28371 deletion (B). the analysis was performed using Nextstrain (https://nextstrain.org/ncov/gisaid/global), based on sequences from the GISAID database (www.epicov.org/epi3/frontend#5c1e82). The lineage in which these mutations occur is circulated with a highlighted circle.
Global phylogenetic analysis shows the uniqueness of the spike 22194-22196 deletion (A) and the nucleocapsid (N) 28362-28371 deletion (B). the analysis was performed using Nextstrain (https://nextstrain.org/ncov/gisaid/global), based on sequences from the GISAID database (www.epicov.org/epi3/frontend#5c1e82). The lineage in which these mutations occur is circulated with a highlighted circle.
Positive Ndel reaction
The positive Ndel reaction is caused by the annealing of the probe to the N deletion at 28362-28371 in the reference sequence. Therefore, the signal will only arise if the deletion is present.
Development of S22194 indel reaction
With this reaction, the Omicron insertion at 22207-22216 and deletion at 22194-22196 are targeted using a probe that binds only to the Omicron sequence containing this pattern,and not to other strains. It was tested along with the inclusive E-sarbeco reaction, including the Delta samples as well. This reaction showed clear specificity with clear identification of Omicron samples.
NSP6del reaction
With this reaction, the primer anneals to the deletion 11283-11292, such that if the deletion is present, the results will be equivalent to those of the E-sarbeco reaction. If not, the signal is inhibited 104-fold due to the absence of primer annealing.
Though the NSPindel mutation is common to the Alpha, Beta, and Gamma VOCs, it was attributed solely to the Delta variant in Israel since that was the only circulating variant in Israel at that time. Moreover, this deletion was found at only 0.03% in of over 3,000 sequences from Israel over the last 3 months. Thus, it was considered to be specific enough for the purpose of identifying the Omicron VOC as compared to the 1% of sequences that showed the presence of the spike gene 69-70 deletion.
When a suspected Omicron sample, along with Alpha and Delta samples, were tested, the assays showed positive Ndel for both Alpha and Omicron, negative or weakly positive Ndel for Delta, and a Cq difference of 12-14 cycles between the E and nsp6del reactions.
As expected, both the Alpha and Omicron-suspected samples were positive for the presence of the NSP6 deletion. The Delta samples were either negative for the NSP6del reaction or weakly positive, with Cq difference of 12-14 cycles between the E reaction and the NSP6del reaction.
Implications
The four assays reported in this paper are ideal for rapid diagnostic workup of a suspected case to identify the presence of the Omicron variant in a simple and cost-effective manner compared to genome sequencing. They can make up part of a scalable rapid protocol with the selected mutations providing highly accurate identification of this VOC.
Since the assays were developed based on the results of testing a small number of positive samples that had been shown to be Omicron by sequencing, their publication at this early stage could be justified only by the urgent demand for a way to distinguish this variant from other circulating strains so as to trace its spread and virulence. This is important in deciding policy measures such as travel restrictions and business closures, which could have potentially devastating economic consequences.
The contribution of the GISAID database was essential in allowing these assays to be validated on a preliminary level. Further testing using other reaction kits with required temperature adjustments will be necessary; however, the researchers expect that the results should be in agreement with this study.
“We plan to conduct a far more thorough and comprehensive validation for these assays and publish our results in a proceeding report. We also welcome and comments and suggestions for improvements, should any such be available, from fellow laboratories.”

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.